MBIC plot visualization
mbicPlot.RdIntuitive way of visualizing how modified BIC values changes across different number of clusters
mbicPlot(seu, criteria="MBIC")Arguments
- seu
an object of class
Seuratrevised by DR.SC with argumentK=NULL.- criteria
a string specifying the information criteria such as AIC, BIC and MBIC to be plotted, default as MBIC.
Details
Nothing
Value
return a ggplot2 object.
References
None
Note
nothing
See also
None
Examples
## we generate the spatial transcriptomics data with lattice neighborhood, i.e. ST platform.
library(Seurat)
data(seu)
seu <- NormalizeData(seu)
#> Normalizing layer: counts
# choose spatially variable features
seu <- FindSVGs(seu)
#> Find the spatially variables genes by SPARK-X...
#> ## ===== SPARK-X INPUT INFORMATION ====
#> ## number of total samples: 100
#> ## number of total genes: 50
#> ## Running with single core, may take some time
#> ## Testing With Projection Kernel
#> ## Testing With Gaussian Kernel 1
#> ## Testing With Gaussian Kernel 2
#> ## Testing With Gaussian Kernel 3
#> ## Testing With Gaussian Kernel 4
#> ## Testing With Gaussian Kernel 5
#> ## Testing With Cosine Kernel 1
#> ## Testing With Cosine Kernel 2
#> ## Testing With Cosine Kernel 3
#> ## Testing With Cosine Kernel 4
#> ## Testing With Cosine Kernel 5
## Just for illustrating the usage of mbicPlot
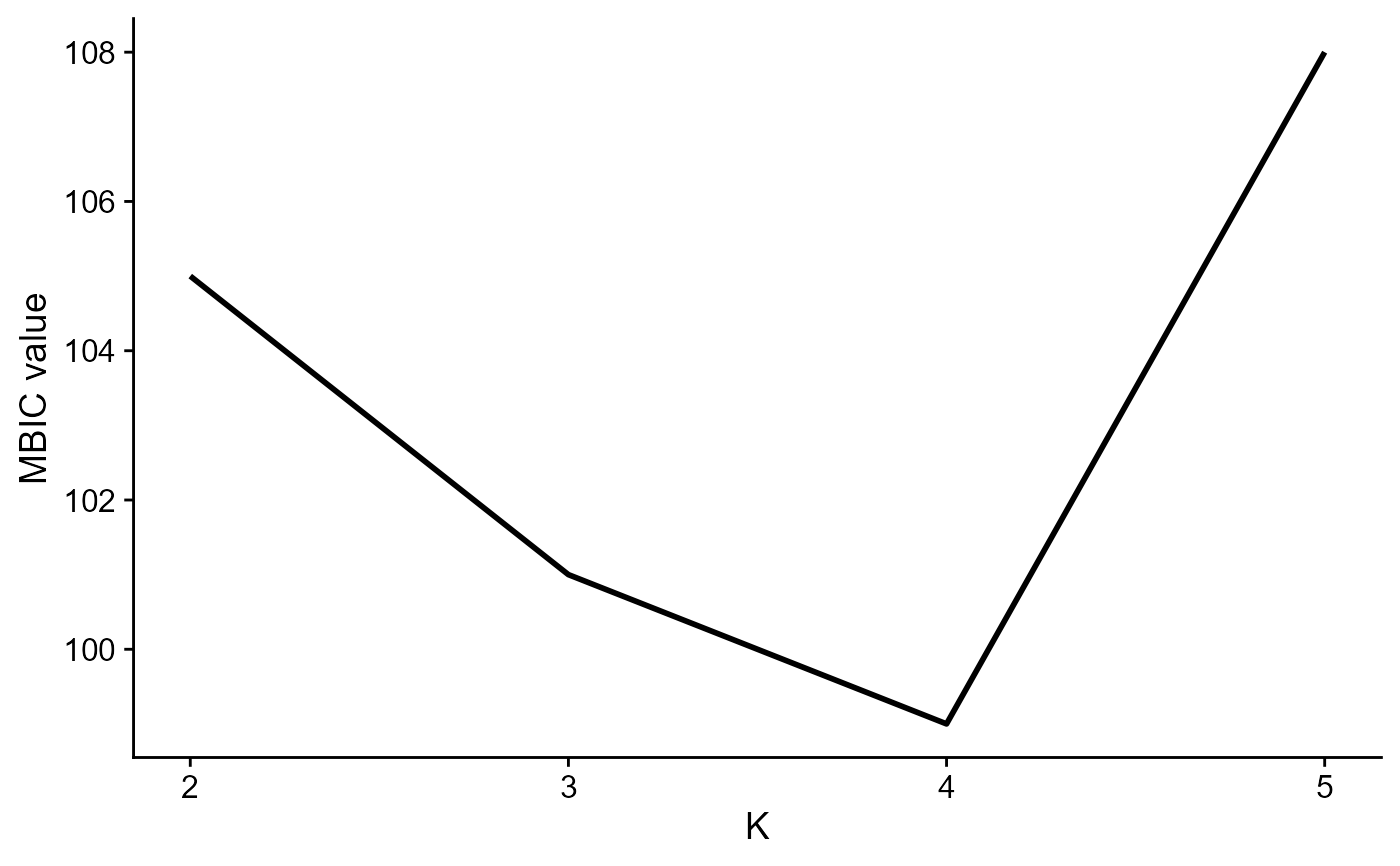
seu[["RNA"]]@misc[['icMat']] <- data.frame(K=2:5, MBIC=c(105, 101, 99, 108))
mbicPlot(seu)