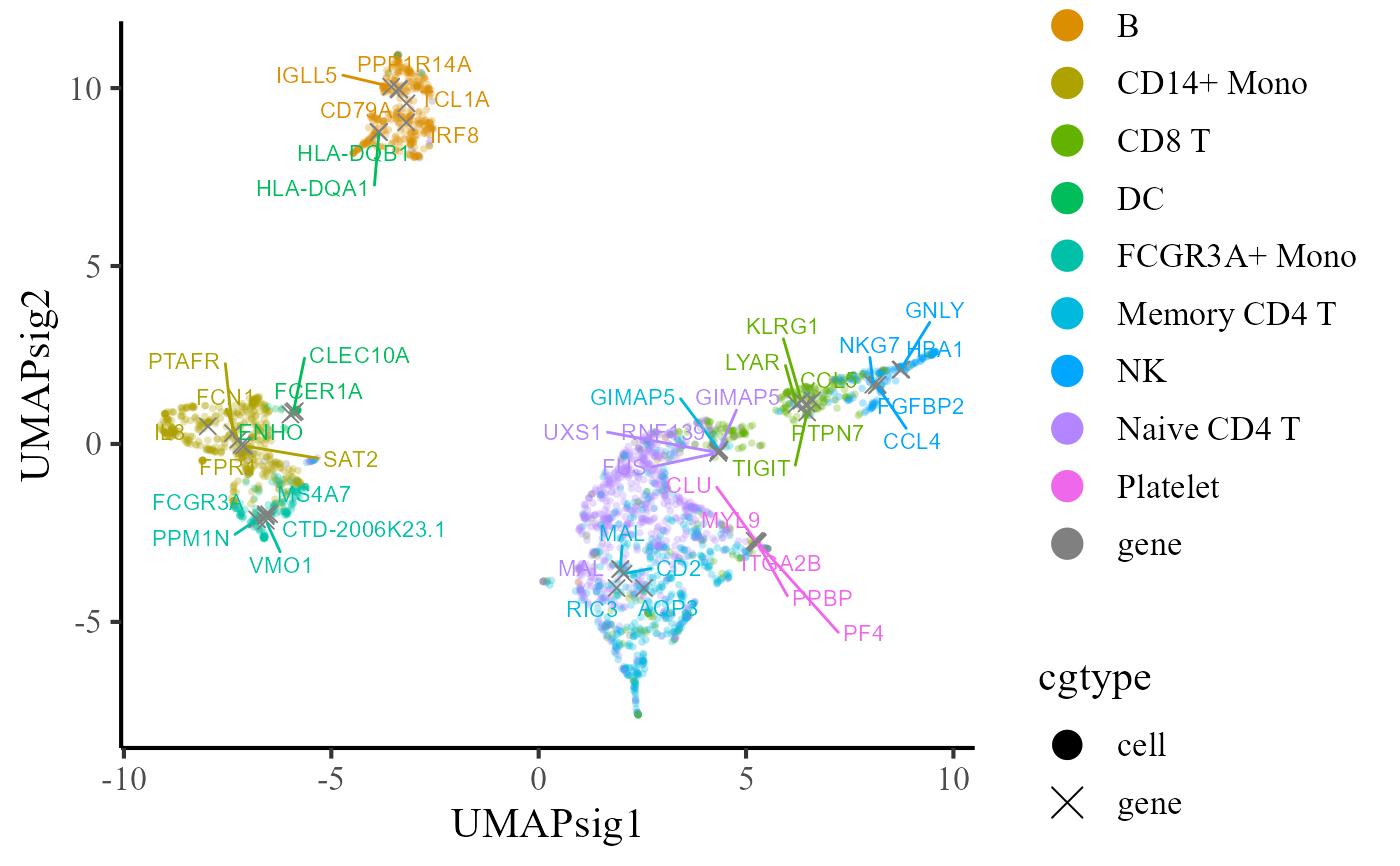
Graph output of a dimensional reduction technique on a 2D scatter plot where each point is a cell or feature and it's positioned based on the coembeddings determined by the reduction technique. By default, cells and their signature features are colored by their identity class (can be changed with the group.by parameter).
Arguments
- seu
a Seurat object with coembedding in the reductions slot wiht component name reduction.
- reduction
a string, specify the reduction component that denotes coembedding.
- gene_txtdata
a data.frame object with columns indcluding `gene` and `label`, specify the cell type/spatial domain and signature genes. Default as NULL, all features will be used in comebeddings.
- cell_label
an optional character in columns of metadata, specify the group of cells/spots. Default as NULL, use Idents as the group.
- xy_name
an optional character, specify the names of x and y-axis, default as the same as reduction.
- dims
a postive integer vector with length 2, specify the two components for visualization.
- cols
an optional string vector, specify the colors for cell group in visualization.
- shape_cg
a positive integers with length 2, specify the shapes of cell/spot and feature in plot.
- pt_size
an optional integer, specify the point size, default as 1.
- pt_text_size
an optional integer, specify the point size of text, default as 5.
- base_size
an optional integer, specify the basic size.
- base_family
an optional character, specify the font.
- legend.point.size
an optional integer, specify the point size of legend.
- legend.key.size
an optional integer, specify the size of legend key.
- alpha
an optional positive real, range from 0 to 1, specify the transparancy of points.
Value
return a ggplot object
Details
None
References
None