GFM: alternate maximization and information criterion
Wei Liu
2023-08-10
Source:vignettes/GFM.Simu.Rmd
GFM.Simu.RmdIn this tutorial, we show that the alternate maximization (AM) is used in the first step of the two-step estimation method and the information criterion (IC) method is adopted to choose the number of factors.
Fit GFM model using simulated data
The package can be loaded with the command:
library("GFM")
#> Loading required package: doSNOW
#> Loading required package: foreach
#> Loading required package: iterators
#> Loading required package: snow
#> Loading required package: parallel
#>
#> Attaching package: 'parallel'
#> The following objects are masked from 'package:snow':
#>
#> clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
#> clusterExport, clusterMap, clusterSplit, makeCluster, parApply,
#> parCapply, parLapply, parRapply, parSapply, splitIndices,
#> stopCluster
#> GFM : Generalized factor model is implemented for ultra-high dimensional data with mixed-type variables.
#> Two algorithms, variational EM and alternate maximization, are designed to implement the generalized factor model,
#> respectively. The factor matrix and loading matrix together with the number of factors can be well estimated.
#> This model can be employed in social and behavioral sciences, economy and finance, and genomics,
#> to extract interpretable nonlinear factors. More details can be referred to
#> Wei Liu, Huazhen Lin, Shurong Zheng and Jin Liu. (2021) <doi:10.1080/01621459.2021.1999818>. Check out our Package website (https://feiyoung.github.io/GFM/docs/index.html) for a more complete description of the methods and analyses
set.seed(1) # set a random seed for reproducibility.GFM can handle data with homogeneous normal variables
First, we generate the data with homogeneous normal variables.
## Homogeneous normal variables
dat <- gendata(q = 2, n=100, p=100, rho=3)Then, we set the algorithm parameters and fit model
# Obtain the observed data
XList <- dat$XList # this is the data in the form of matrix list.
str(XList)
#> List of 1
#> $ : num [1:100, 1:100] -1.031 1.528 -0.565 1.288 -0.667 ...
#> ..- attr(*, "dimnames")=List of 2
#> .. ..$ : NULL
#> .. ..$ : NULL
X <- dat$X # this is the data in form of matrix
# set variables' type, 'gaussian' means there is continous variable type.
types <- 'gaussian' Third, we fit the GFM model with user-specified number of factors.
# specify q=2
gfm1 <- gfm(XList, types, algorithm="AM", q=2, verbose = FALSE)
# measure the performance of GFM estimators in terms of canonical correlations
measurefun(gfm1$hH, dat$H0, type='ccor')
#> [1] 0.9893027
measurefun(gfm1$hB, dat$B0, type='ccor')
#> [1] 0.9931255The number of factors can also be determined by data-driven manners.
# select q automatically
hq <- chooseFacNumber(XList, types, select_method='IC', q_set = 1:6, verbose = FALSE, parallelList=list(parallel=TRUE))
#>
|
| | 0%
|
|============== | 20%
|
|============================ | 40%
|
|========================================== | 60%
|
|======================================================== | 80%
|
|======================================================================| 100%
#> IC criterion estimates the factor number q as 2.
hq
#> [1] 2GFM outperforms LFM in analyzing data with heterogeous normal variables
First, we generate the data with heterogeous normal variables and set the parameters of algorithm.
dat <- gendata(seed=1, n=100, p=100, type='heternorm', q=2, rho=1)
# Obtain the observed data
XList <- dat$XList # this is the data in the form of matrix list.
str(XList)
#> List of 1
#> $ : num [1:100, 1:100] 0.7458 -2.006 1.705 0.0484 3.094 ...
#> ..- attr(*, "dimnames")=List of 2
#> .. ..$ : NULL
#> .. ..$ : NULL
X <- dat$X # this is the data in form of matrix
# set variables' type, 'gaussian' means there is continous variable type.
types <- 'gaussian' Third, we fit the GFM model with user-specified number of factors and compare the results with that of linear factor models.
# specify q=2
gfm1 <- gfm(XList, types, algorithm="AM", q=2, verbose = FALSE)
# measure the performance of GFM estimators in terms of canonical correlations
corH_gfm <- measurefun(gfm1$hH, dat$H0, type='ccor')
corB_gfm <- measurefun(gfm1$hB, dat$B0, type='ccor')
lfm1 <- Factorm(X, q=2)
corH_lfm <- measurefun(lfm1$hH, dat$H0, type='ccor')
corB_lfm <- measurefun(lfm1$hB, dat$B0, type='ccor')
library(ggplot2)
#> Warning: package 'ggplot2' was built under R version 4.1.3
df1 <- data.frame(CCor= c(corH_gfm, corH_lfm, corB_gfm, corB_lfm),
Method =factor(rep(c('GFM', "LFM"), times=2)),
Quantity= factor(c(rep('factors',2), rep("loadings", 2))))
ggplot(data=df1, aes(x=Quantity, y=CCor, fill=Method)) + geom_bar(position = "dodge", stat="identity",width = 0.5)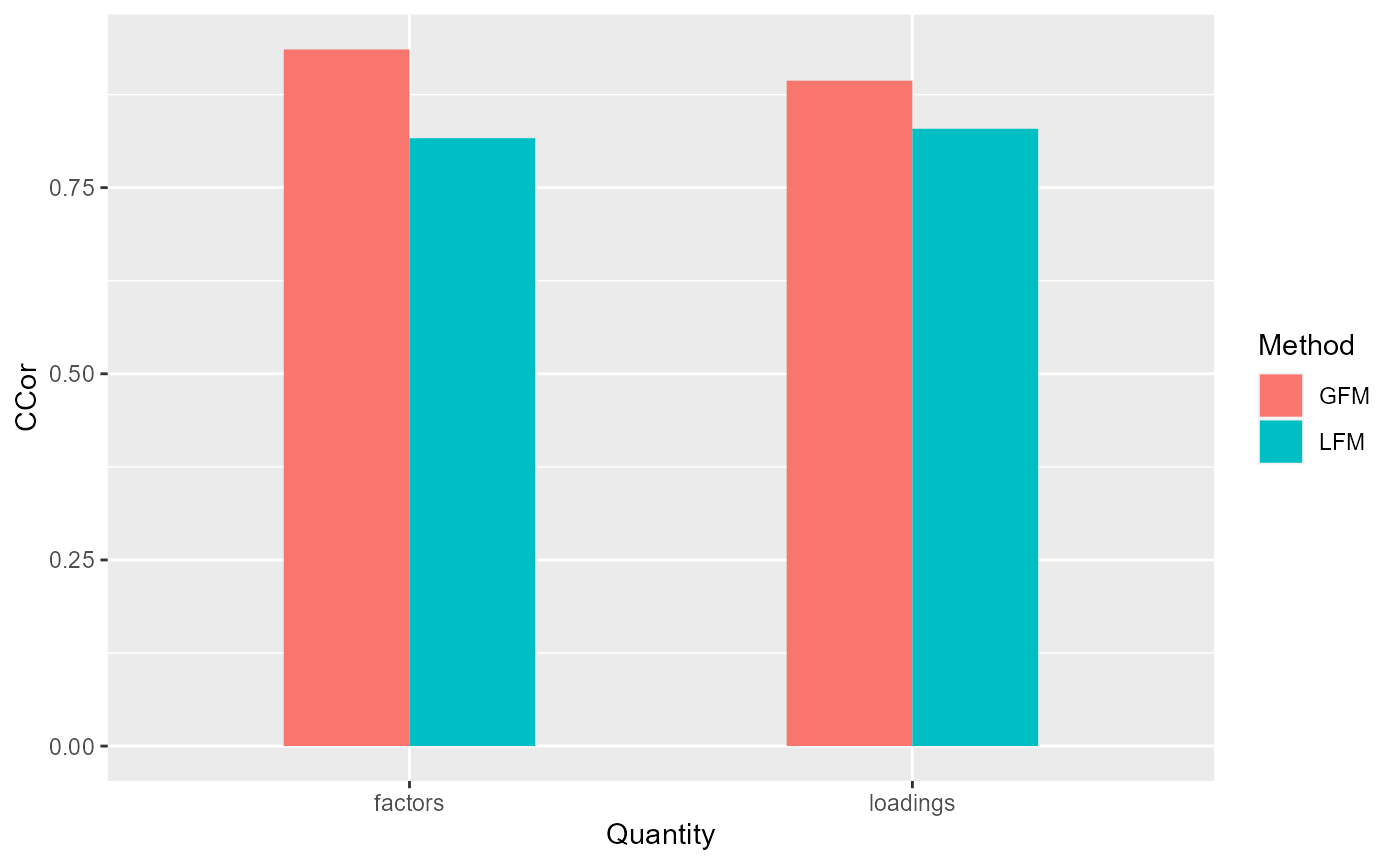
The number of factors can also be determined by data-driven manners.
# select q automatically
hq <- chooseFacNumber(XList, types, select_method='IC', q_set = 1:6, verbose = FALSE, parallelList=list(parallel=TRUE))
#>
|
| | 0%
|
|============== | 20%
|
|============================ | 40%
|
|========================================== | 60%
|
|======================================================== | 80%
|
|======================================================================| 100%
#> IC criterion estimates the factor number q as 2.GFM outperforms LFM in analyzing data with Count(Poisson) variables
First, we generate the data with Count(Poisson) variables and set the parameters of algorithm.
q <- 3; p <- 200
dat <- gendata(seed=1, n=200, p=p, type='pois', q=q, rho=4)
# Obtain the observed data
XList <- dat$XList # this is the data in the form of matrix list.
str(XList)
#> List of 1
#> $ : int [1:200, 1:200] 0 3 4 1 0 4 2 3 1 1 ...
X <- dat$X # this is the data in form of matrix
# set variables' type, 'gaussian' means there is continous variable type.
types <- 'poisson'Second, we we fit the GFM models given the true number of factors.
system.time(
gfm1 <- gfm(XList, types, algorithm="AM", q=3, verbose = FALSE)
)
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> user system elapsed
#> 4.36 0.03 9.40system.time(
hq <- chooseFacNumber(XList, types, q_set=1:6, select_method = "IC", parallelList=list(parallel=TRUE))
)
#>
|
| | 0%
|
|============== | 20%
|
|============================ | 40%
|
|========================================== | 60%
|
|======================================================== | 80%
|
|======================================================================| 100%
#> IC criterion estimates the factor number q as 3.
#> user system elapsed
#> 0.00 0.02 12.83Third, we compare the results with that of linear factor models.
# measure the performance of GFM estimators in terms of canonical correlations
corH_gfm <- measurefun(gfm1$hH, dat$H0, type='ccor')
corB_gfm <- measurefun(gfm1$hB, dat$B0, type='ccor')
lfm1 <- Factorm(X, q=3)
corH_lfm <- measurefun(lfm1$hH, dat$H0, type='ccor')
corB_lfm <- measurefun(lfm1$hB, dat$B0, type='ccor')
library(ggplot2)
df1 <- data.frame(CCor= c(corH_gfm, corH_lfm, corB_gfm, corB_lfm),
Method =factor(rep(c('GFM', "LFM"), times=2)),
Quantity= factor(c(rep('factors',2), rep("loadings", 2))))
ggplot(data=df1, aes(x=Quantity, y=CCor, fill=Method)) + geom_bar(position = "dodge", stat="identity",width = 0.5)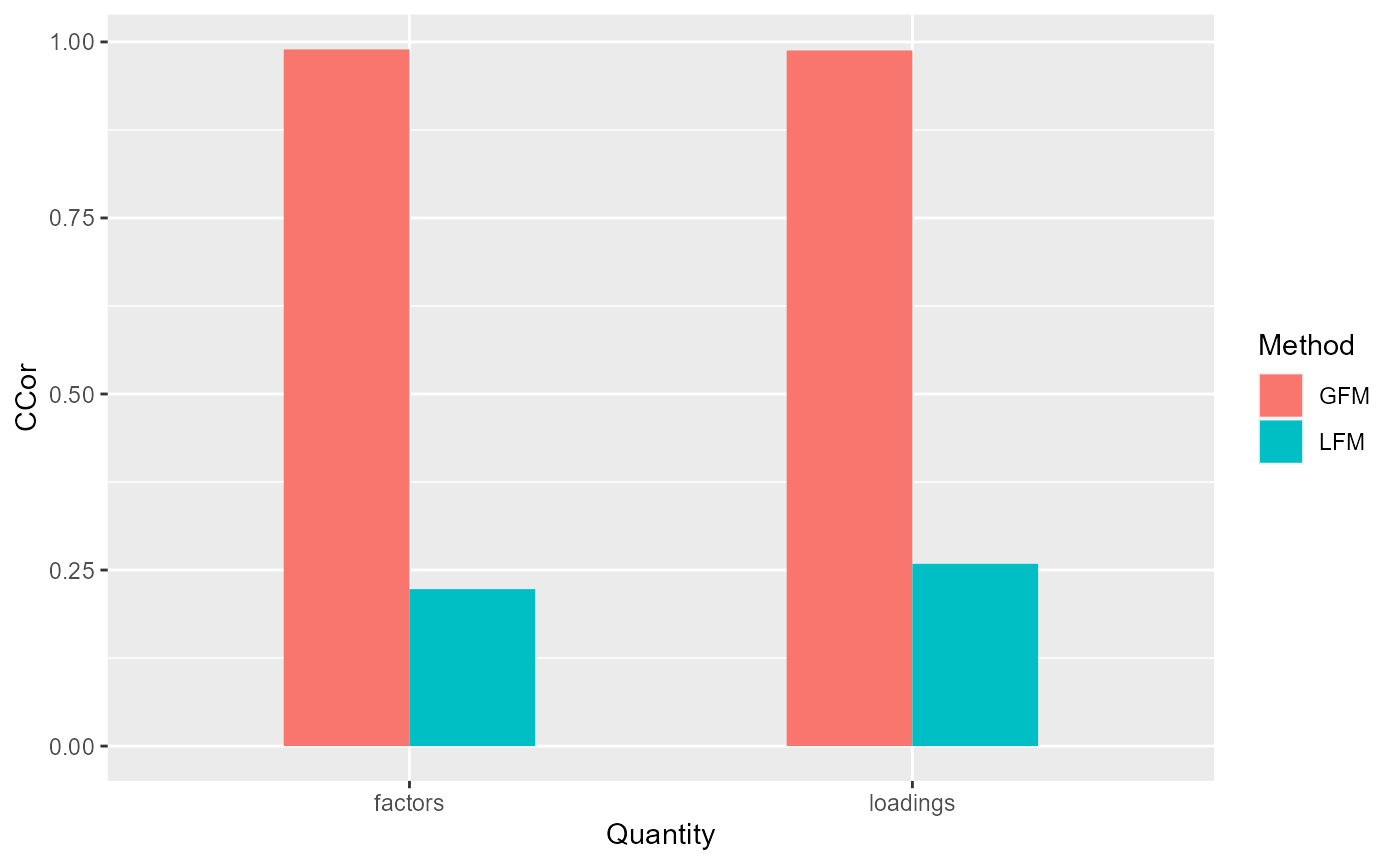
GFM outperforms LFM in analyzing data with the mixed-types of count and categorical variables
First, we generate the data with Count(Poisson) variables and set the parameters of algorithm. Then fit the GFM model with user-specified number of factors.
dat <- gendata(seed=1, n=200, p=200, type='pois_bino', q=2, rho=2)
# Obtain the observed data
XList <- dat$XList # this is the data in the form of matrix list.
str(XList)
#> List of 2
#> $ : int [1:200, 1:100] 2 0 3 5 1 1 1 5 1 0 ...
#> $ : int [1:200, 1:100] 1 0 1 1 0 0 1 0 0 0 ...
X <- dat$X # this is the data in form of matrix
# set variables' type, 'gaussian' means there is continous variable type.
types <- dat$types
table(dat$X[,1])
#>
#> 0 1 2 3 4 5 6 8 11
#> 52 61 34 26 14 8 3 1 1
table(dat$X[, 200])
#>
#> 0 1
#> 108 92
# user-specified q=2
gfm2 <- gfm(XList, types, algorithm="AM", q=2, verbose = FALSE)
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
#> Warning: glm.fit: fitted rates numerically 0 occurred
measurefun(gfm2$hH, dat$H0, type='ccor')
#> [1] 0.9820777
measurefun(gfm2$hB, dat$B0, type='ccor')
#> [1] 0.960852Third, we compare the results with that of linear factor models.
# select q automatically
hq <- chooseFacNumber(XList, types, select_method='IC', q_set = 1:4, verbose = FALSE, parallelList=list(parallel=TRUE))
#>
|
| | 0%
|
|======================= | 33%
|
|=============================================== | 67%
|
|======================================================================| 100%
#> IC criterion estimates the factor number q as 2.
# measure the performance of GFM estimators in terms of canonical correlations
corH_gfm <- measurefun(gfm2$hH, dat$H0, type='ccor')
corB_gfm <- measurefun(gfm2$hB, dat$B0, type='ccor')
Compare with linear factor models
lfm1 <- Factorm(dat$X, q=3)
corH_lfm <- measurefun(lfm1$hH, dat$H0, type='ccor')
corB_lfm <- measurefun(lfm1$hB, dat$B0, type='ccor')
library(ggplot2)
df1 <- data.frame(CCor= c(corH_gfm, corH_lfm, corB_gfm, corB_lfm),
Method =factor(rep(c('GFM', "LFM"), times=2)),
Quantity= factor(c(rep('factors',2), rep("loadings", 2))))
ggplot(data=df1, aes(x=Quantity, y=CCor, fill=Method)) + geom_bar(position = "dodge", stat="identity",width = 0.5)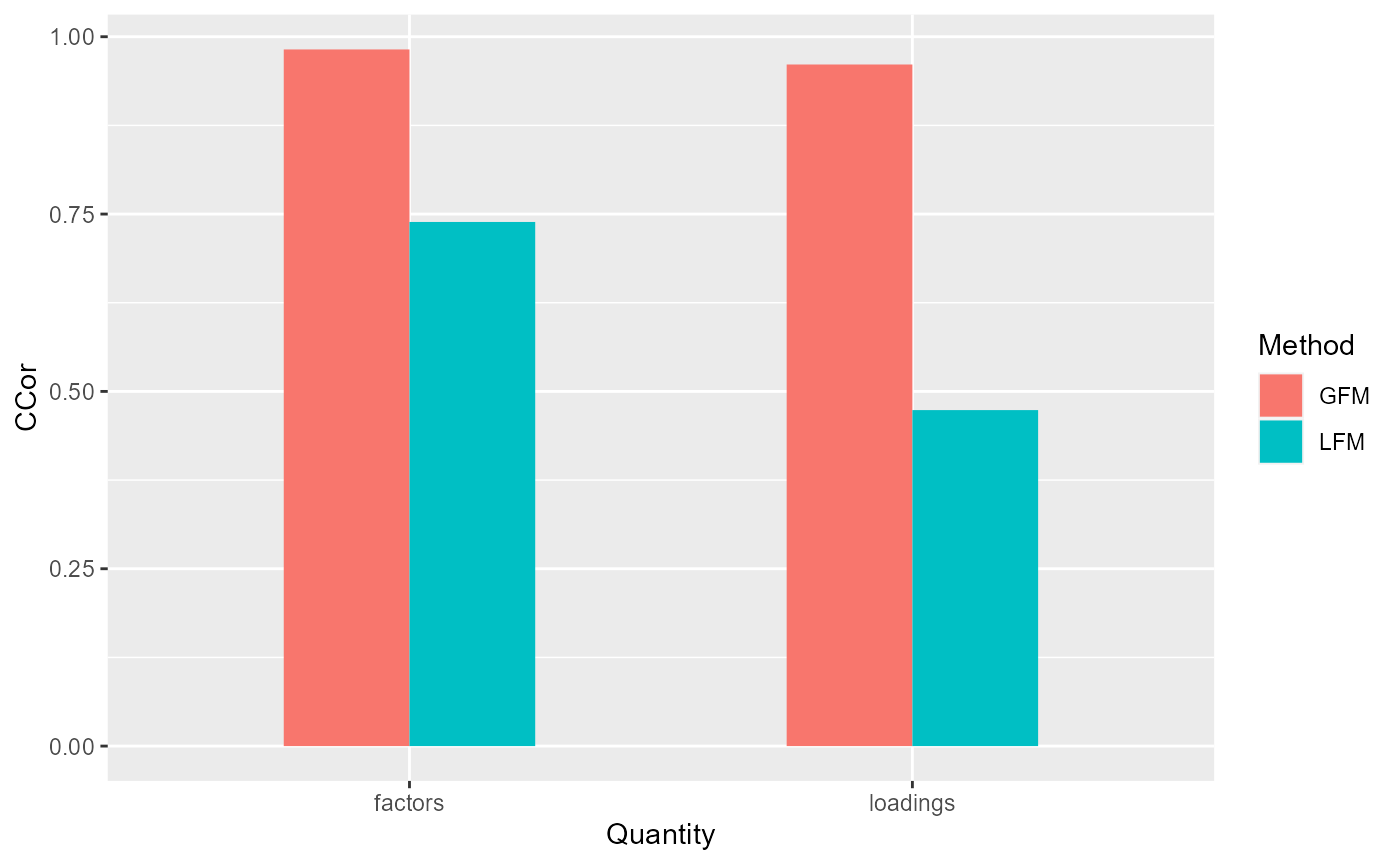
Session information
sessionInfo()
#> R version 4.1.2 (2021-11-01)
#> Platform: x86_64-w64-mingw32/x64 (64-bit)
#> Running under: Windows 10 x64 (build 22621)
#>
#> Matrix products: default
#>
#> locale:
#> [1] LC_COLLATE=Chinese (Simplified)_China.936
#> [2] LC_CTYPE=Chinese (Simplified)_China.936
#> [3] LC_MONETARY=Chinese (Simplified)_China.936
#> [4] LC_NUMERIC=C
#> [5] LC_TIME=Chinese (Simplified)_China.936
#>
#> attached base packages:
#> [1] parallel stats graphics grDevices utils datasets methods
#> [8] base
#>
#> other attached packages:
#> [1] ggplot2_3.4.1 GFM_1.2.1 doSNOW_1.0.20 snow_0.4-4
#> [5] iterators_1.0.14 foreach_1.5.2
#>
#> loaded via a namespace (and not attached):
#> [1] tidyselect_1.1.2 xfun_0.29 bslib_0.3.1 purrr_0.3.4
#> [5] lattice_0.20-45 generics_0.1.2 colorspace_2.1-0 vctrs_0.6.1
#> [9] htmltools_0.5.2 yaml_2.3.6 utf8_1.2.3 rlang_1.1.0
#> [13] pkgdown_2.0.6 jquerylib_0.1.4 pillar_1.9.0 withr_2.5.0
#> [17] DBI_1.1.2 glue_1.6.2 lifecycle_1.0.3 stringr_1.4.0
#> [21] munsell_0.5.0 gtable_0.3.3 ragg_1.2.2 codetools_0.2-18
#> [25] memoise_2.0.1 evaluate_0.15 labeling_0.4.2 knitr_1.37
#> [29] fastmap_1.1.0 irlba_2.3.5 fansi_1.0.4 highr_0.9
#> [33] Rcpp_1.0.10 scales_1.2.1 cachem_1.0.6 desc_1.4.0
#> [37] jsonlite_1.8.0 farver_2.1.1 systemfonts_1.0.4 fs_1.5.2
#> [41] textshaping_0.3.6 digest_0.6.29 stringi_1.7.6 dplyr_1.0.9
#> [45] rprojroot_2.0.3 grid_4.1.2 cli_3.2.0 tools_4.1.2
#> [49] magrittr_2.0.3 sass_0.4.1 tibble_3.2.1 crayon_1.5.1
#> [53] pkgconfig_2.0.3 MASS_7.3-55 Matrix_1.4-0 assertthat_0.2.1
#> [57] rmarkdown_2.11 rstudioapi_0.13 R6_2.5.1 compiler_4.1.2